KGGA(Knowledge-based Genetic and Genomic Analyses Accelerator)
Introduction
KGGA is an open-source, high-performance computational platform engineered to revolutionize large-scale genomic studies. Leveraging the GTB genomic compression framework and CCF distributed computing architecture, KGGA achieves >10–100X acceleration in processing cohorts exceeding 1 million subjects while maintaining analytical rigor, enabling population-scale genomics on standard workstations (≥8 cores, 16GB RAM). By democratizing supercomputer-grade genomics for standard research environments, KGGA delivers an industrial-grade solution for large-scale GWAS, rare variant analysis, and clinical genomic interpretation.
Functionality
KGGA currently supports the following function modules:
clean: a basic module that makes genotype-level and variant-level QC, and extracts variant and sample subgroups. The processed variants can be exported into various formats (e.g., plink, pgen, and VCF) or further analyzed by the other functional modules on KGGA.annotate: a module that quickly annotates variants using hundreds of fields from multiple databases simultaneously, and filters variants based on the annotations.prune: a module that prunes variants according to association p-value, linkage disequilibrium (LD), and annotation scores. The genotypes of pruned variants can be exported in multiple formats for follow-up analyses.prioritize: a module that prioritizes variants, genes, or genomic regions based on allelic frequencies in reference populations by various comprehensive analysis pipelines.connect: designed for advanced users, KGGA also provides a suite of high-performance API functions that enable direct interaction with third-party applications built using Java, Python, or R. These APIs support bidirectional integration, allowing them to be seamlessly invoked by external tools as well.
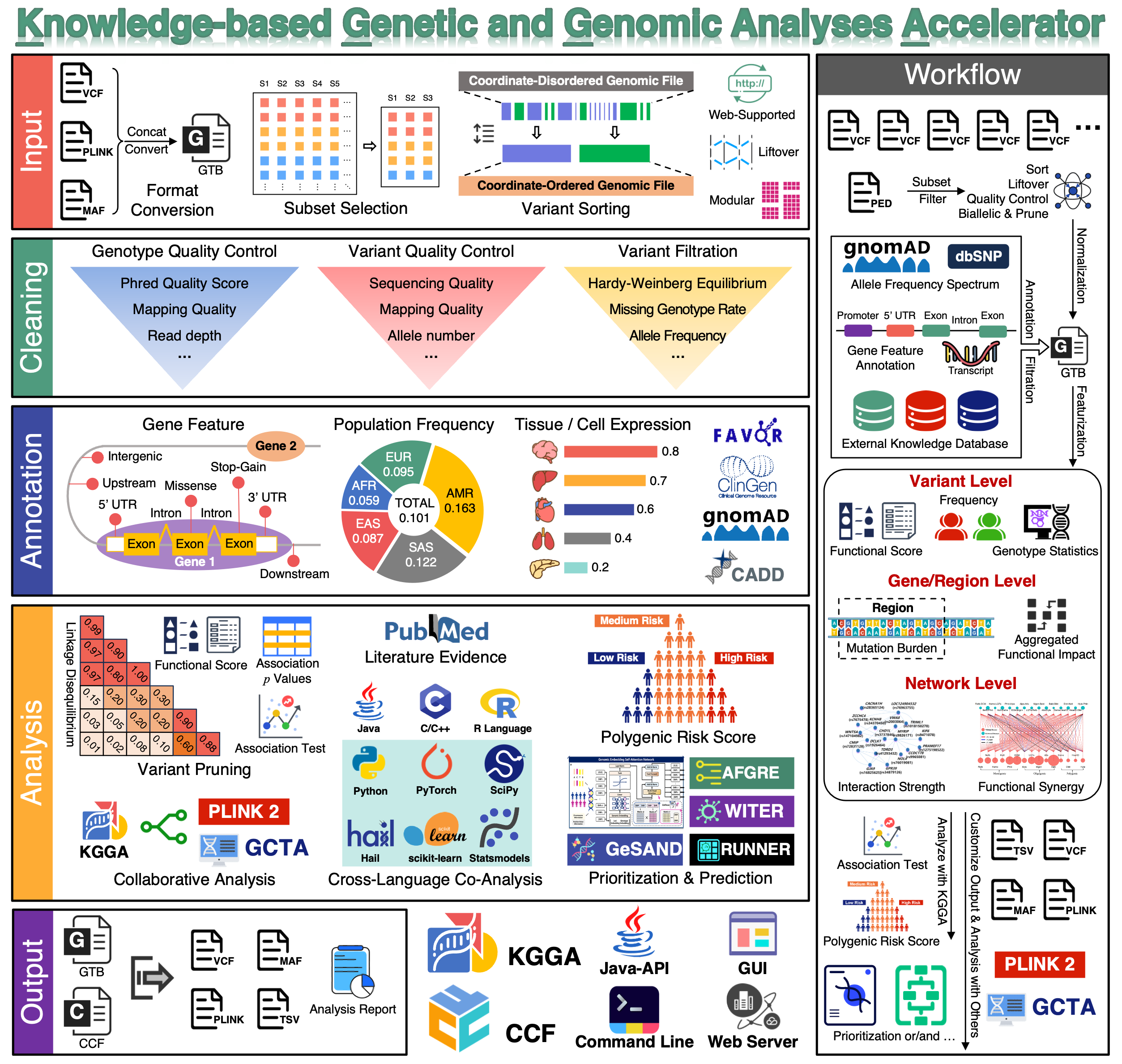
Key Advances of KGGA
- Ultra-high speed and minimal memory footprint, approaching hardware limits
- Modular incremental execution & resumable workflows
- Extensive biomedical knowledge base
- Seamless integration with analysis tools and platforms via R APIs or binary data
Citations
The GTB format paper: Zhang L, Yuan Y, Peng W, Tang B, Li MJ, Gui H, Wang Q, Li M*. GBC: A parallel toolkit based on fast-accessible byte blocks for extremely large-scale genotypes of species. Genome Biol. 2023 Apr 17;24(1):76.