[!TIP|label:This is an archived version]
GBC-stable-1.0 version has been released and can be accessed at http://pmglab.top/gbc.
About GBC
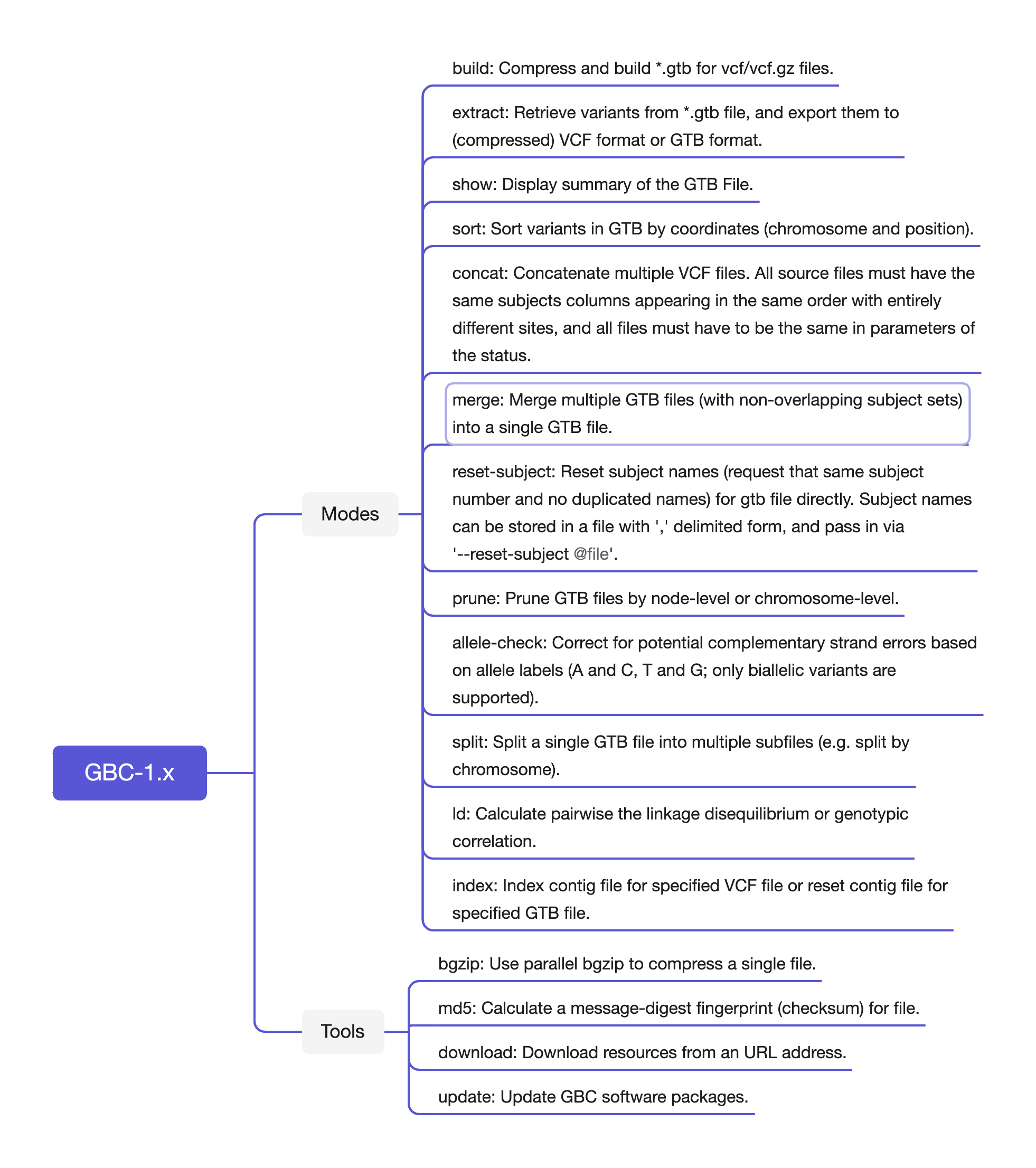
GBC (short for GenoType Blocking Compressor) is a blocking compressor for genotype data, which aims at creating a unified and flexible structure-GenoType Block (GTB) for genotype data in the variant call format (VCF) files. There will be a less occupation of hard disk space, a faster data access and extraction function, a more convenient management of population files and a more efficient precess of data analysis with the GTB structure compared with the conventional gz format. GBC provides the following functions:
Efficient compression: Linearly increasing time overhead with sample and variant size, stable memory utilization and competitive compression ratio.
- Memory: Single-threaded compression with < 4 GB memory usage.
- Speed: up to 78516269 genotypes/s.
Quality control: Quality control at variant-level, genotype-level, population-allele frequency/count-level. The open API facilitates users to customize their own filtering methods.
- Quick query: query continuous/random variants, filter variants by allele frequency/count, extract subset of samples, etc;
- File management: merge, join, split, subset sample selection, sorting, allele label checking, etc;
- Complex calculations: LD calculations.
- Genotype coding for a wide range of haploid/diploid species.
GBC is a free standalone toolkit for improved the efficiency of storage and file management of large-scale genotypes. GBC is also a library of fundamental API development tools that can be easily integrated into existing tool flows to accelerate the analysis and computation process of genotype-based data.
[!COMMENT|label:Contact Developer]
Liubin Zhang, suranyi.sysu@gmail.com