Variant Annotation & Filtration
About
The annotate module in KGGA is designed to assist with genetic variant analysis, a critical process in understanding genetic variations and their impacts on health and disease. This module provides a flexible and comprehensive approach to annotating and filtering variants, making it easier for researchers to focus on relevant genetic data from large datasets.
What the Module Does
The annotate module enables the rapid annotation of millions of genetic variants using one or multiple databases. It allows users to select specific fields or entire databases for annotation, leveraging tools like gnomAD for allele frequency, dbNSFP and CADD for prediction scores, and others for expression and gene feature data. This is particularly useful for large-scale genomic studies where efficiency and accuracy are paramount.
Workflow Analysis
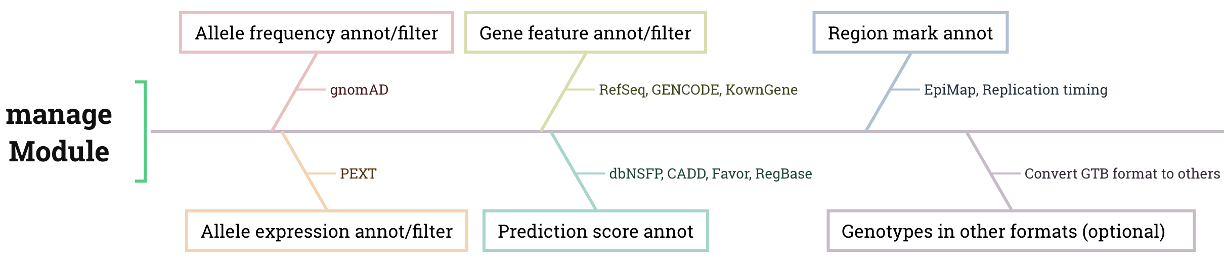
The workflow is described as follows, following the manage module in KGGA:
- Allele Frequency Annotation & Filtration
- Annotation: This step involves adding population-level allele frequency information, such as minor allele frequency, often sourced from databases like gnomAD. For example, it may include data on how common a variant is in specific populations.
- Filtration: Variants are then selected based on specific frequency criteria, such as those with a minor allele frequency below a threshold in particular population ancestries. This is crucial for identifying rare variants potentially linked to diseases.
- Allele Expression Annotation & Filtration
- Annotation: This adds expression level information, such as PEXT (Proportion Expression Across Transcripts) scores from gnomAD, which summarize exonic region expression across tissues based on GTEx data (gnomAD PEXT). This helps assess how variants might affect gene expression.
- Filtration: Variants are filtered based on expression levels, such as selecting those with high expression in certain tissues, which can be relevant for studying tissue-specific effects.
- Gene Feature Annotation & Filtration
- Annotation: This determines which gene or transcript a variant affects and classifies its functional role, such as missense (changing an amino acid) or nonsense (introducing a premature stop codon). This is standard in tools like ANNOVAR or VEP, used for genomic location and impact assessment.
- Filtration: Variants are selected based on specific gene features, such as those in coding regions, particular genes, or functional categories, aiding in prioritizing variants for further analysis.
- Prediction Score Annotation
- Annotation: This step adds prediction scores from databases like dbNSFP, CADD, and FAVOR, which provide functional or pathogenic impact assessments. For instance, CADD scores predict variant deleteriousness, while dbNSFP offers multiple prediction algorithms. The module is noted for allowing rapid, one-stop annotation of hundreds of fields from one or multiple databases, enhancing efficiency for large-scale studies.
- Notably, this step does not mention filtration, suggesting it may be used in subsequent analyses rather than filtered within this module.
- Region Mark Annotation
- Annotation: This involves adding information from databases with genomic interval features, such as epigenomic markers (e.g., histone modifications, DNase hypersensitivity sites), to highlight variants that regulate gene expression. This is important for understanding non-coding variant effects, often overlooked in traditional analyses.
- Like prediction scores, this step lacks a mentioned filtration process, possibly indicating its role is primarily annotative for later use.
Output and Format
After processing, the module saves the filtered variants in a custom GTB (Genotype Block) binary format, which is efficient for further analysis in other KGGA modules. It also outputs results in text format for human readability, ensuring accessibility for researchers.
Basic Usage
java -jar kgga.jar annotate --input <input1> --input <input2> --output <output> [options]
The annotate module in KGGA is designed to streamline the process of variant quality control and gene feature annotation by default. However, if you do not require these processes, the module offers the flexibility to disable them, allowing you to tailor the annotation workflow to your specific needs.