Variant & Sample Cleaning
About
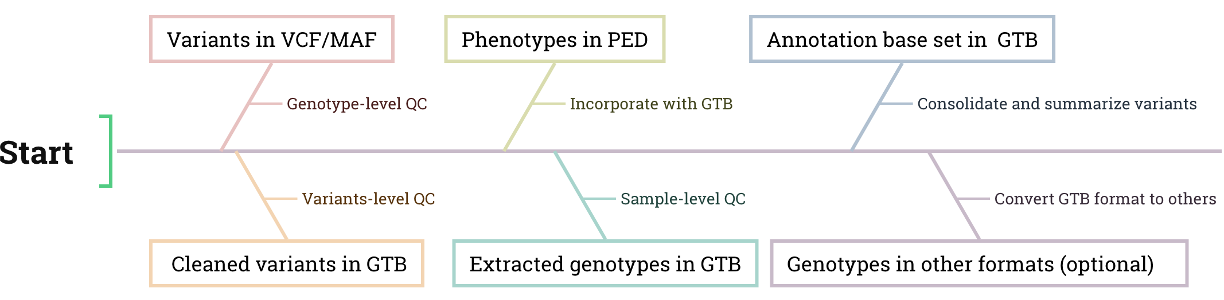
The clean Module is the only way that must be passed for all other Modules of KGGA. It produces a high-quality subset of variants and extracts genotypes of samples in the efficient binary format GTB for follow-up analyses.
Workflow of the Manage Module
Genotype-level Quality Control: Removing genotypes with low sequencing quality, low read depth, or confusion.
Variant-level Quality Control: Removing variants with low sequencing quality, low mapping quality, or other filtration criteria (e.g., allele number, allele frequency, specific INFO field of VCF).
The above two steps are based on the VCF file(s) only.
Incorporate Phenotype Data: Integrating phenotype data of subjects from the PED file to extract their corresponding genotypes.
Sample-level Variant Selection: Selecting variants based on mutation type, allele frequency, missing genotype rate, or the Hardy-Weinberg equilibrium test in a specific subset of samples.
Generate Annotation Base Set: Consolidate all cleaned variants from input files (where applicable) into a unified GTB-format variant file. This dataset retains only genomic coordinates (hg38/GRCh38), alleles, and genotype frequency summaries, forming the foundational dataset for downstream KGGA analyses.
Note: In the last two steps, if a PED file is provided, the genotype data for individuals who are present in both the VCF and PED files will be utilized for the selection process. Without a PED file, the selection will proceed based solely on the genotype data available for all individuals listed in the VCF file.
Basic Usage
java -jar kgga.jar clean --input <input1> --input <input2> --output <output> [options]
If no other option is set, the program will perform some preset filters by default. Users are also allowed to set custom filtration criteria if needed or disable QC entirely.