Variant Annotation & Filtration
About
The annotate module enables rapid annotation of millions of variants with genomic features using one or multiple different databases, leveraging the full or partial fields of these databases. Additionally, it offers a variety of filtering functions based on annotation results, such as gene feature filtering, population frequency, conservation, and epigenetic modification, to assist in interpreting and deciphering the significant association signals.
Workflow of the Annotate Module
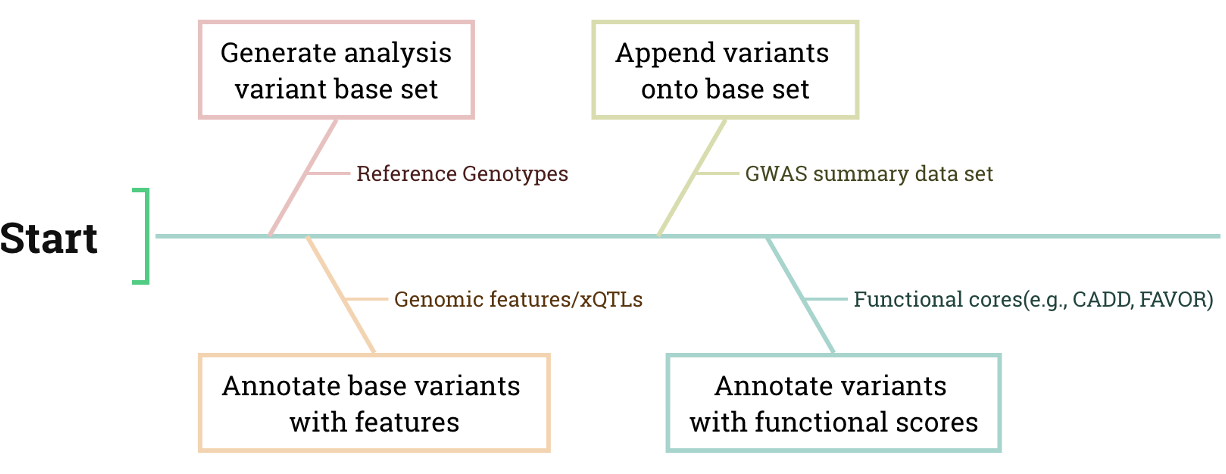
Generation: Extract variant coordinates and frequencies from the VCF or GBC file to create a root variant set for further analysis.
Gene Annotation: Annotate the root variant with gene features or xQTLs.
Append: Integrate GWAS variants and their summary statistics into the annotated root variant set.
Variant Annotation: Annotate variants with databases (e.g., CADD - Combined Annotation Dependent Depletion, and gnomAD) stored in GTB format to gain comprehensive insights into your genetic variations. KGGSum allows for rapid, one-stop annotation of hundreds of fields from one or multiple databases.
Clump and Prune: Clump and prune GWAS variants using p-values from summary statistics and LD calculated from a reference population.
Basic Usage
java -jar kggsum.jar annot --sum-file <input1> --ref-gty-file <input2> --output <output> [options]