Association
About
The association module in KGGSum offers various functions to link genes, cell types, gene networks, and even drugs to phenotypes using GWAS signals from variants. A common step across all analyses involves aggregating association signals from multiple variants to derive gene-level associations using GATES (Li et al., 2011) and ECS (Li et al., 2019). Advanced association analyses build upon these gene-level associations. The rationale behind each type of association analysis is detailed in the respective option descriptions and relevant publications.
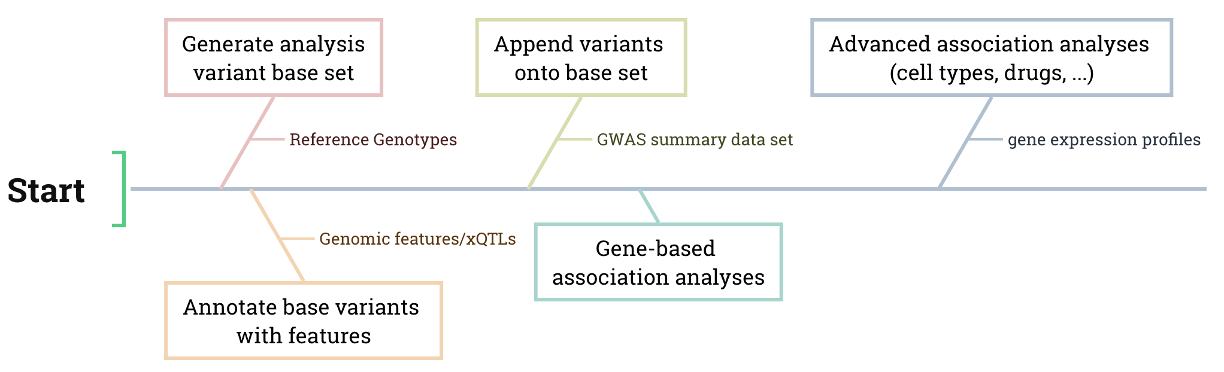
Main Workflow of the Association Module
Generation: Extract variant coordinates and frequencies from the VCF or GBC file to create a root variant set for further analysis.
Annotation: Annotate the root variant with gene features or xQTLs.
- Append: Integrate GWAS variants and their summary statistics into the annotated root variant set.
- Gene-based association: Conduct gene-based association analyses based on the gene feature annotations.
- Advanced association: Conduct gene-based association analyses based on the gene feature annotations.
- …: (Additional association analysis as specified).
Basic Usage
java -jar kggsum.jar assoc --ref-gty-file <input1> --sum-file <input2> --output <output> [options]
By default, the program performs gene-based association tests using GATES and ECS. Additional association analyses in this module can be initiated by specifying relevant input options (e.g., --gene-score-file).