Causation inference
About
The causation module provides advanced Mendelian randomization methods for inferring causation from genes, lifestyle, or microbes (as exposures) to phenotypes (as outcomes). It enables rapid inference screening of tens of exposures and outcomes simultaneously.
Main Workflow of the Causation Module
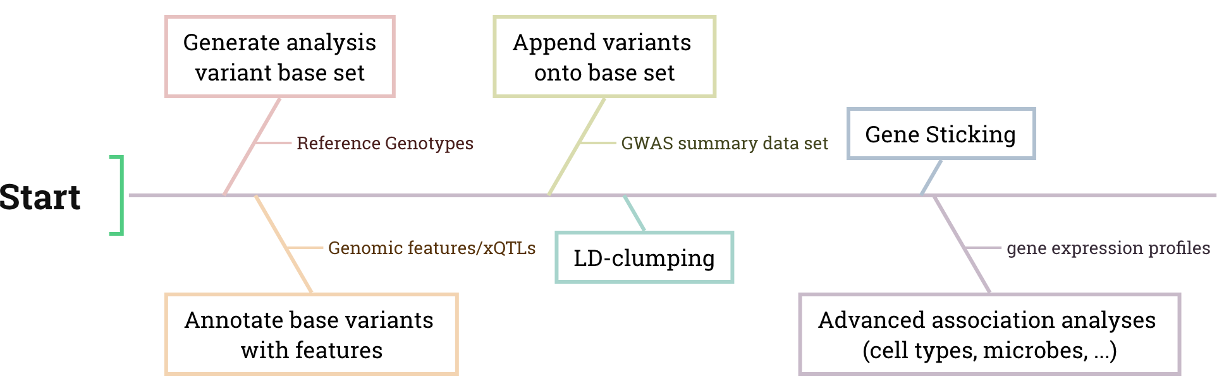
Generation: Extract variant coordinates and frequencies from the VCF or GBC file to create a root variant set for further analysis.
Annotation: Annotate the root variant with gene features or xQTLs.
- Append: Integrate GWAS variants and their summary statistics into the annotated root variant set.
- LD Clumping: Select the significant variants of exposures as IVs and remove redundant IVs according to LD.
- Gene Sticking: Link IV to the potential target gene according to LD. Note that this is a rough linking, and some target genes may not be true.
- MR analysis: Infer the causality by suitable MR methods (e.g., EMIC or PCMR)
- …: (Additional analysis as specified).
Basic Usage
java -jar kggsum.jar causal --sum-file <input1> --ref-gty-file <input2> --output <output> [options]