About HttpDownloader
Researches on bioinformatics and medical informatics require a large amount of public resources. However, due to their huge data volume (e.g., dbNSFP database reaches over 30 GB, gnomAD database reaches over 1 TB), users are always suffering from many problems (e.g., redirection jump, speed limit in the cloud, breakpoint transfer, etc.) when downloading these resources. In order to simplify the process of downloading and updating these databases, and integrate this process as an API tool into other public projects (mainly for Java development) effectively, we have developed the HttpDownloader. In addition, support for FTP downloads is being planned.
HttpDownloader is a simple Http downloader based on the Java platform, designed to enhance the download function for basic resources. HttpDownloader now supports for: breakpoint transfer, URL redirection based on the Location of the response header, parallel download (requires the server to support chunked or get 206 response codes after sending segmented requests), dynamically getting file size for resource downloads, proxy setting.
API
The main API methods for HttpDownloader can be found in edu.sysu.pmglab.downloader.HttpDownloader, which has been integrated in the commandParser package, including:
- download: HttpDownloader.instance(String url)
- Set the number of threads for parallel download: .setThreads(int nThreads)
- Set the output file name: .setOutputFile(File outputFile) 和 .setOutputFile(String outputFileName)
- Set the temporary path for cache data: .setTempDir(File tempDir) 和 .setTempDir(File tempDirName)
- Set the proxy: .setProxy(String host, String port) 和 .setProxy(String hostPort)
- Set the maximum waitting time (unit: second): .setTimeOut(int timeOut)
- Clearing cached data: .clean(boolean clean)
When the instantiation task is complete, submit the corresponding task by .download().
Design Parser
The first parameter of input command will be identified as the URL address by HttpDownloader, and the following parameters will be parsed as options (set offset=1). The steps of parser creation are described as following:
Step1: Create command group:Options;
Step2: Create command items for Options group: --output; --temp-dir, --threads, --overwrite, --proxy, --time-out, --no-auto-retry;
Step3: Set the program name: <mode>, set offset as 1;
Step4: Set the Usage Style as Unix_Style_3 and set the subtitle as follows:
Step5: Export to Java Script Builder With Options Format.
Design Main Function
Use CommandParser to bridge input parameters with business logic. See: HttpDownloader.java.
public static void main(String[] args) {
try {
if (args.length == 0) {
System.out.println(HttpDownloaderParser.getParser());
return;
}
HttpDownloaderParser parser = HttpDownloaderParser.parse(args);
if (parser.help.isPassedIn) {
System.out.println(HttpDownloaderParser.getParser());
return;
}
do {
try {
HttpDownloader.instance(args[0])
.setOutputFile(parser.output.value)
.setThreads(parser.threads.value)
.setPrintLog(true)
.setTempDir(parser.tempDir.value)
.setProxy(parser.proxy.value)
.setTimeOut(parser.timeout.value)
.clean(parser.overwrite.isPassedIn)
.download();
} catch (IOException e) {
logger.error("{}", e.getMessage());
if (!parser.noAutoRetry.isPassedIn) {
logger.error("Download failed, resume download in 3 seconds.");
Thread.sleep(3000);
continue;
}
}
break;
} while (true);
} catch (Exception | Error e) {
logger.error("{}", e.getMessage());
}
}
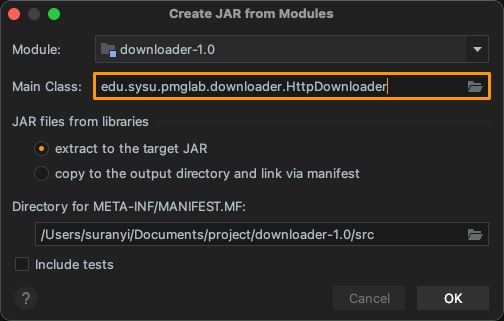
Create Jar Package
Click: "Project Structure..." > "Artifacts" > "+" > "JAR" > "From modules with dependencies...". Select the entry function in Main Class, and packaged as downloader-1.0.jar:
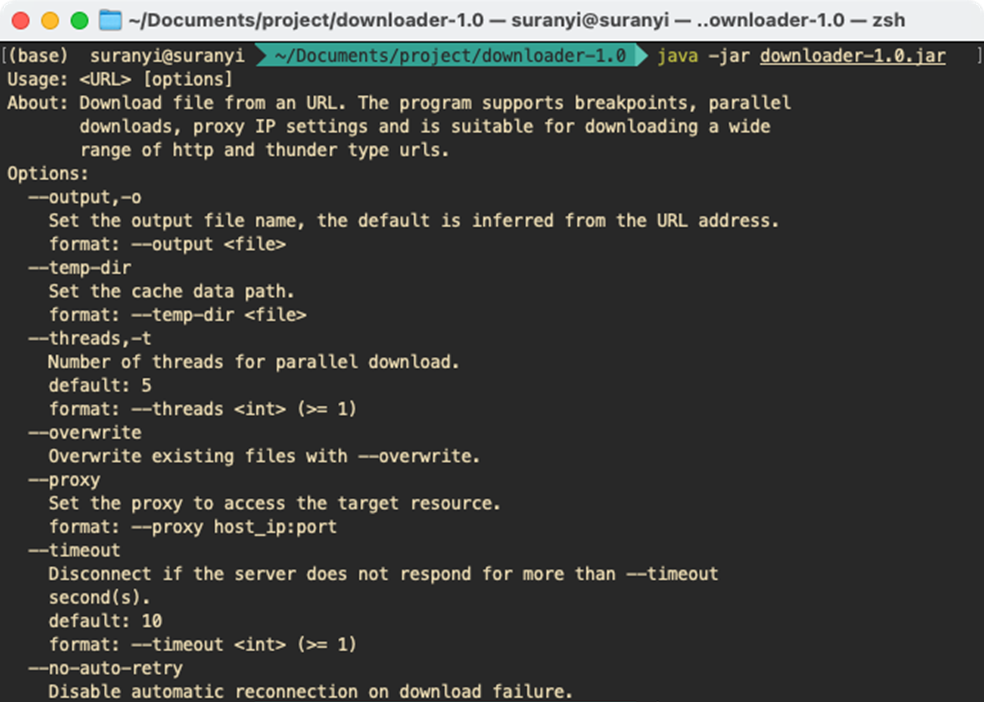
Go to the folder where downloader-1.0.jar is located, and enter the command on the console to display the document:
java -jar downloader-1.0.jar
Example
1. Download gnomAD
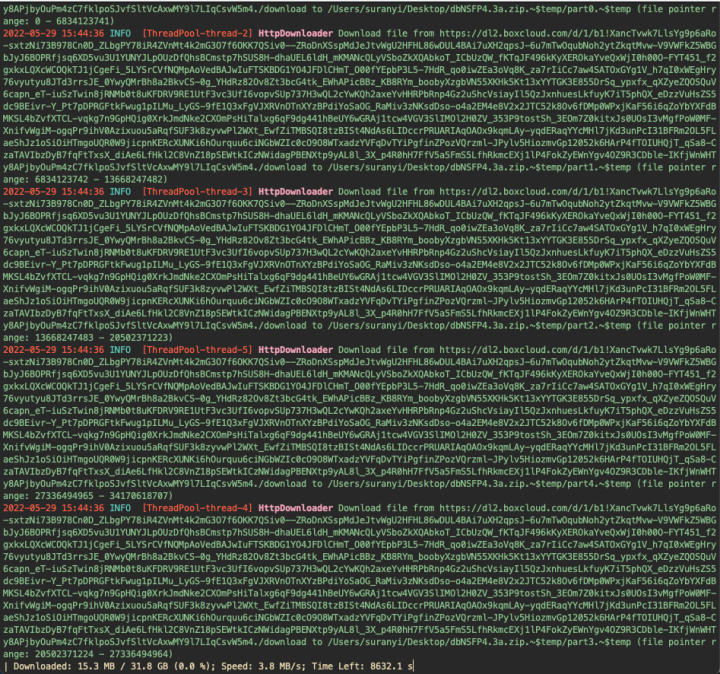
The Genome Aggregation Database (gnomeAD) is a collaborative genomic mutation frequency database created by national researchers to aggregate and coordinate large-scale sequencing projects at different levels, including whole-exome and whole-genome data, for a wide range of scientific research communities. The database currently includes 125,748 whole-exome data and 15,708 whole-genome data (v2) from different disease research projects and large population sequencing projects. The database includes the previously used 1,000 genomes data, the ESP database and most of the ExAC database.
Amazon's links support parallel downloading and resumable without the need for a proxy.
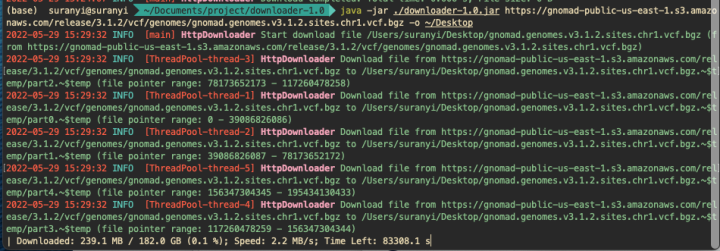
java -jar ./downloader-1.0.jar https://gnomad-public-us-east-1.s3.amazonaws.com/release/3.1.2/vcf/genomes/gnomad.genomes.v3.1.2.sites.chr1.vcf.bgz -o ~/Desktop
2. Download dbNSFP
dbNSFP is a database developed for functional prediction and annotation of all potential non-synonymous single-nucleotide variants (nsSNVs) in the human genome. Its current version is based on the Gencode release 29 / Ensembl version 94 and includes a total of 84,013,490 nsSNVs and ssSNVs (splicing-site SNVs). It compiles prediction scores from 38 prediction algorithms (SIFT, SIFT4G, Polyphen2-HDIV, Polyphen2-HVAR, LRT, MutationTaster2, MutationAssessor, FATHMM, MetaSVM, MetaLR, MetaRNN, CADD, CADD_hg19, VEST4, PROVEAN, FATHMM-MKL coding, FATHMM-XF coding, fitCons x 4, LINSIGHT, DANN, GenoCanyon, Eigen, Eigen-PC, M-CAP, REVEL, MutPred, MVP, MPC, PrimateAI, GEOGEN2, BayesDel_addAF, BayesDel_noAF, ClinPred, LIST-S2, ALoFT), 9 conservation scores (PhyloP x 3, phastCons x 3, GERP++, SiPhy and bStatistic) and other related information including allele frequencies observed in the 1000 Genomes Project phase 3 data, UK10K cohorts data, ExAC consortium data, gnomAD data and the NHLBI Exome Sequencing Project ESP6500 data, various gene IDs from different databases, functional descriptions of genes, gene expression and gene interaction information, etc.
java -jar ./downloader-1.0.jar https://dl2.boxcloud.com/d/1/b1\!XancTvwk7LlsYg9p6aRo-sxtzNi73B978Cn0D_ZLbgPY78iR4ZVnMt4k2mG3O7f6OKK7QSiv0--ZRoDnXSspMdJeJtvWgU2HFHL86wDUL4BAi7uXH2qpsJ-6u7mTwOqubNoh2ytZkqtMvw-V9VWFkZ5WBGbJyJ6BOPRfjsq6XD5vu3U1YUNYJLpOUzDfQhsBCmstp7hSUS8H-dhaUEL6ldH_mKMANcQLyVSboZkXQAbkoT_ICbUzQW_fKTqJF496kKyXEROkaYveQxWjI0h00O-FYT451_f2gxkxLQXcWCOQkTJ1jCgeFi_5LYSrCVfNQMpAoVedBAJwIuFTSKBDG1YO4JFDlCHmT_O00fYEpbP3L5-7HdR_qo0iwZEa3oVq8K_za7rIiCc7aw4SATOxGYg1V_h7qI0xWEgHry76vyutyu8JTd3rrsJE_0YwyQMrBh8a2BkvCS-0g_YHdRz82Ov8Zt3bcG4tk_EWhAPicBBz_KB8RYm_boobyXzgbVN55XKHk5Kt13xYYTGK3E855DrSq_ypxfx_qXZyeZQOSQuV6capn_eT-iuSzTwin8jRNMb0t8uKFDRV9RE1UtF3vc3UfI6vopvSUp737H3wQL2cYwKQh2axeYvHHRPbRnp4Gz2uShcVsiayIl5QzJxnhuesLkfuyK7iT5phQX_eDzzVuHsZS5dc9BEivr-Y_Pt7pDPRGFtkFwug1pILMu_LyGS-9fE1Q3xFgVJXRVnOTnXYzBPdiYoSaOG_RaMiv3zNKsdDso-o4a2EM4e8V2x2JTC52k8Ov6fDMp0WPxjKaF56i6qZoYbYXFdBMKSL4bZvfXTCL-vqkg7n9GpHQig0XrkJmdNke2CXOmPsHiTalxg6qF9dg441hBeUY6wGRAj1tcw4VGV3SlIMOl2H0ZV_353P9tostSh_3EOm7Z0kitxJs0UOsI3vMgfPoW0MF-XnifvWgiM-ogqPr9ihV0Azixuou5aRqfSUF3k8zyvwPl2WXt_EwfZiTMBSQI8tzBISt4NdAs6LIDccrPRUARIAqOAOx9kqmLAy-yqdERaqYYcMHl7jKd3unPcI31BFRm2OL5FLaeShJz1oSiOiHTmgoUQR0W9jicpnKERcXUNKi6hOurquu6ciNGbWZIc0cO9O8WTxadzYVFqDvTYiPgfinZPozVQrzml-JPylv5HiozmvGp12052k6HArP4fTOIUHQjT_qSa8-CzaTAVIbzDyB7fqFtTxsX_diAe6LfHkl2C8VnZ18pSEWtkICzNWidagPBENXtp9yAL8l_3X_p4R0hH7FfV5a5FmS5LfhRkmcEXj1lP4FokZyEWnYgv4OZ9R3CDble-IKfjWnWHTy8APjbyOuPm4zC7fklpoSJvfSltVcAxwMY9l7LIqCsvW5m4./download -o ~/Desktop/dbNSFP4.3a.zip