About GBC
The GenoType Block Compressor (GBC) is an assembly of tools for compressing, accessing and managing genotype data with unparalleled efficiency. At its core is the GenoType Block (GTB), a novel data structure optimized for swift addressing and minimal storage of ultra-large datasets.
Through parallel algorithms, GBC achieves lightningspeed data compression, access, and analysis. It replaces traditional formats with GTB to significantly reduce storage requirements while accelerating cohort management and populationscale computation.
GBC encompasses GTB and high-performance IO frameworks. GTB enables efficient management of massive datasets. Optimized parallel procedures allow GBC to realize rapid data access and management. By substituting traditional formats with GTB, GBC decreases storage needs, hastens data handling, and facilitates analysis.
[!TIP|label:Citation|style:callout]
Zhang, L., Yuan, Y., Peng, W. et al. GBC: a parallel toolkit based on highly addressable byte-encoding blocks for extremely large-scale genotypes of species. Genome Biol 24, 76 (2023). https://doi.org/10.1186/s13059-023-02906-z
Why do we need GBC?
- For ordinary users: you can easily manage and quickly access large-scale (millions of subjects) whole-genome genotype data and annotation data resources, and directly analyze variant and genotype data quickly through tools such as PLINK, KGGSeq.
- For developers: GBC provides a rich and high-performance Java API function library, and Java, Python (JPype), R (rJava), C++ and other languages can build their own efficient analysis of large-scale project processes and tools based on it.
Highlights of GBC
- Efficient compression: GBC boasts outstanding compression ratios, while consuming less than 2 GB of memory with a throughput of up to 25,000,000 genotypes/s when compressing data in a single thread. It also supports using multiple threads to improve compression speed.
- Fast access and query: With GBC, users can easily query continuous/random variants, filter variants by allele frequency/count, filter variants by specified field conditions, and extract subset samples. These features enable rapid access and querying of genotypes.
- Speed up calculation: GBC accelerates the calculation of linkage disequilibrium coefficients between large-scale samples' variants. With IO optimization, GBC expect to effectively accelerate more computational methods.
- Quality control: quality control at the variant level, genotype level, population allele frequency level, and retain extended interfaces.
- File management: GBC facilitates file management through a range of features including merge, connect, split, extract sample subsets, and sort functions.
- Unified encoding of haploid/diploid species multi-allele genotypes: GBC uses a unified and lossless encoding scheme for all genotypes of a variant, enabling easy merging and exchange of genotype data across different species.
- HTTP/HTTPS access: In addition to reading local input data at high speed, GBC also enables direct access to remote data via HTTP/HTTPS, thereby allowing for remote data processing.
- Support for multiple output formats: GBC supports output in various formats, including GTB, VCF, TSV, CSV, PLINK-BED and others. Additionally, users can customize columns (column name, column type, column formatting, etc.) to facilitate file protocol exchange between downstream analyses.
[!NOTE|label:Support for multiple file format conversions|style:callout]
GBC currently supports parallel and direct mutual conversion between GTB format and VCF, MAF, TSV, and BED formats. If you have a format specification that needs to be implemented, please send the implementation details of the algorithm and relevant reference papers to Liubin Zhang suranyi.sysu@gmail.com. We will implement them in subsequent updated versions.
You can also refer to https://pmglab.top/gbc/api-docs/edu/sysu/pmglab/gbc/toolkit/bed/package-summary.html to implement your own file format conversions. We also welcome you to actively send the Java scripts implementing the methods to Liubin Zhang suranyi.sysu@gmail.com. We will include them in updated versions of GBC after testing.
Benchmarks
Compression Performance
We compared GBC's genotypes compression performance with that of BGT (Heng Li, 2016), GTShark (Sebastian Deorowicz, 2019), Genozip (Divon Lan et al., 2020), and BCFTools (Heng Li, 2010) on UKBB-exome-chr1, which contains 469,835 subjects and 1,890,165 variants.
| Software | Compressed Size [GB] | Compression Time [hour] | Decompression Time [hour] |
|---|---|---|---|
| GBC | 11.598 | 3.558 | 2.732 |
| BGT | 32.073 | 18.810 | 8.975 |
| GTShark | 9.886 | 6.230 | 9.005 |
| Genozip | 9.244 | 10.620 | 17.694 |
| BCFTools | 25.682 | 8.990 | 6.467 |
Notes:
- Similar tools that provide fast access such as GTC (Sebastian Deorowicz, 2018) and PBWT (Richard Durbin, 2014) were unable to build a compression archive for this dataset (GTC compression time > 2 months; PBWT memory usage > 100 GB).
- All software compression and decompression were performed using 2 threads (if possible). For tools that do not support parallel decompression to the BGZF format, we processed the VCF text information from standard output using pipes and redirects
| bgzip -c -l 5 > $output.- In this experiment, whole exome genotypes from UK Biobank was accessed through a collaboration with application ID 86920. Data are available for bona fide researchers upon application to the UK Biobank. These genotypes were first processed using the default parameters of GBC (including quality control of genotypes and variants, following the official guidelines and the practice experience of KGGSeq), to remove non-genotype data, and then converted back to the VCF format for testing with all software.
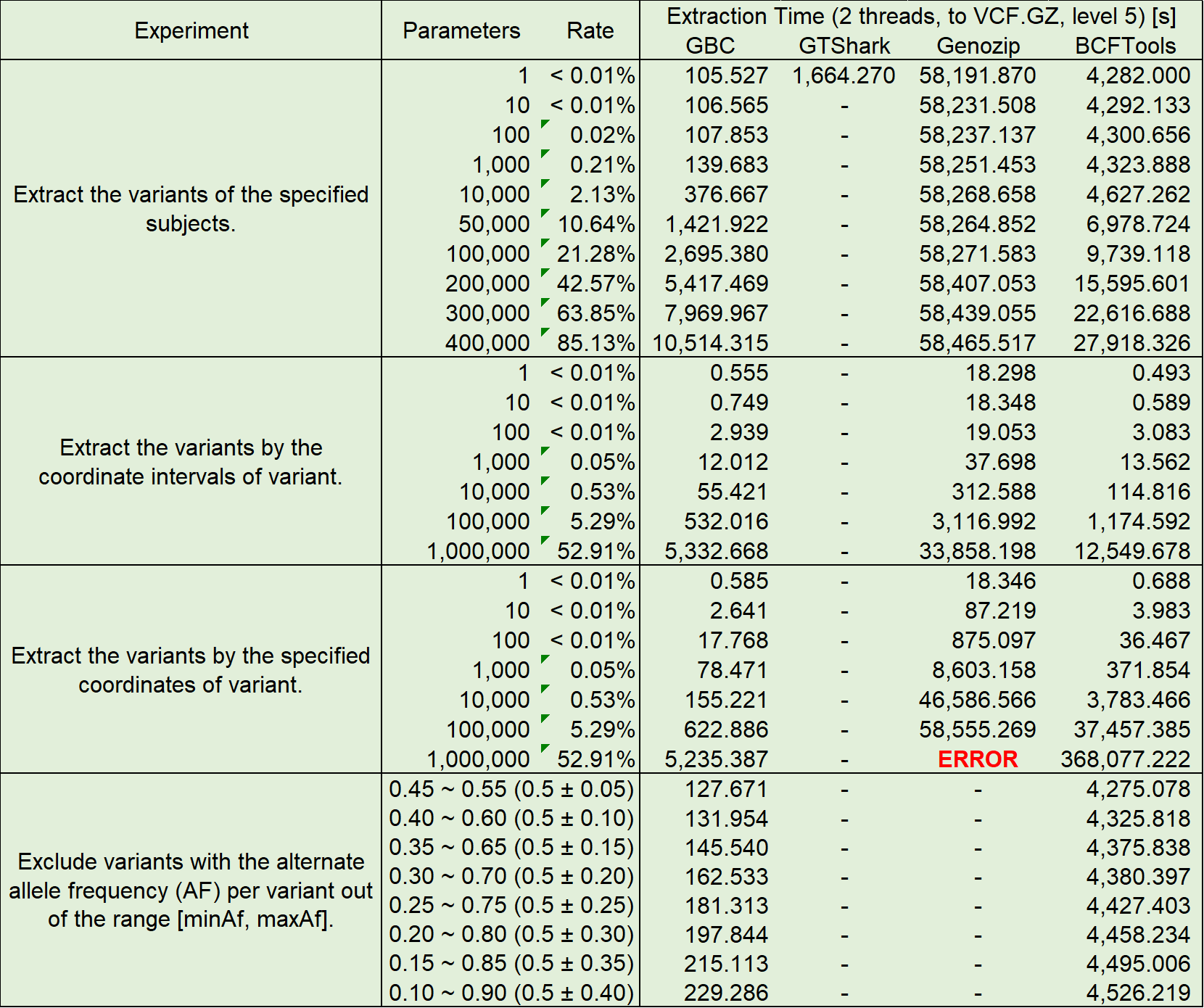
Query Performance
The compressed archives built above were used for four performance tests, the test items and results are shown in the following table: